0. Previous posting
1. Sample data
2. Coxph model
3. Hazard Ratio Table
4. Meta analysis
5. Forestplot
6. Kaplan-Meier
0. Previous posting
- Survival 기본 학습
2023.04.10 - [R] - [R][Survival Analysis] 생존 분석, Kaplan-Meier curve, Cox-hazard, Hazard Ratio
[R][Survival Analysis] 생존 분석, Kaplan-Meier curve, Cox-hazard, Hazard Ratio
1. 생존 분석 이론 2. Tutorial 3. Advanced 1.생존 분석 이론 - From ChatGPT Survival Analysis는 시간에 따른 사건(예: 사망, 질병 발생 등)이 일어날 확률을 계산하는 분석 방법입니다. [2] 이를 위해 전문 회귀
datainsider.tistory.com
1. Sample data
- 샘플 데이터 만들기
library(dplyr)
library(data.table)
set.seed(230706)
# toy data
dt<-data.table(
pid = sample(1:5000, 2000, replace = F),
mace = sample(0:1, 2000, replace = T, prob = c(0.8,0.2)),
mace_sub = sample(c("acute_mi","is","cr","death","cvd_death"), 2000, replace = T, prob = rep(0.2,5)),
dm = sample(0:1, 2000, replace = T, prob = c(0.8,0.2)),
dementia = sample(0:1, 2000, replace = T, prob = c(0.8,0.2)),
myalgia = sample(0:1, 2000, replace = T, prob = c(0.8,0.2)),
age = sample(0:99, 2000, replace = T),
group = sample(c("Atorvastatin","Pravastatin"), 2000, replace = T, prob = c(0.8,0.2)),
from = sample(c("0","1","2","3","4","5","6","7","8"), 2000, replace = T),
index_year = sample(2006:2022, 2000, replace = T),
event_tars_year = sample(0:15, 2000, replace = T)
)
glimpse(dt)
2. Coxph model
- Coxph model function으로 만든 후 결과 보기
library(survival)
create_main_table <- function(data, outcome){
hos_list<-unique(data$from)
raw_table <- data.table()
meta <- data.table()
dt_now <- data.table()
for (i in 1:length(hos_list)) {
hos <- hos_list[i]
dt_now <- data[from==hos]
n_p <- nrow(dt_now[group=="Pravastatin"])
n_p_e <- nrow(dt_now[group=="Pravastatin"&get(outcome)==1])
ir_p <- round(((n_p_e/sum(dt_now[group=="Pravastatin"]$event_tars_year))*1000),2)
n_a <- nrow(dt_now[group=="Atorvastatin"])
n_a_e <- nrow(dt_now[group=="Atorvastatin"&get(outcome)==1])
ir_a <- round(((n_a_e/sum(dt_now[group=="Atorvastatin"]$event_tars_year))*1000),2)
# input variables by institution
dt_now[,(outcome):=as.numeric(get(outcome))]
fit_now <- tryCatch({
coxph(Surv(event_tars_year, get(outcome)) ~ group, data = dt_now)
}, error = function(e) {
NA
})
if (outcome %in% c("myal","myal2")) {
# cat("myal coxph")
fit_now2 <- tryCatch({
coxph(Surv(event_tars_year, get(outcome)) ~ group+index_year+alcohol+Antiplatelet_drug, data = dt_now) #+department_type
}, error = function(e) {
NA
})
}
else {
# cat("not myal coxph")
fit_now2 <- tryCatch({
coxph(Surv(event_tars_year, get(outcome)) ~ group + index_year, data = dt_now) #+department_type
}, error = function(e) {
NA
})
}
HR <- tryCatch({
paste0(round(exp(coef(fit_now)),2),"[",round(exp(confint(fit_now)[1]),2),";",round(exp(confint(fit_now)[2]),2),"]")
}, error = function(e) {
NA
})
p <- tryCatch({
round(summary(fit_now)$coefficients[, "Pr(>|z|)"],3)
}, error = function(e) {
NA
})
HR2 <- tryCatch({
paste0(round(exp(coef(fit_now2))[1],2),"[",round(exp(confint(fit_now2)[1,1]),2),";",round(exp(confint(fit_now2)[1,2]),2),"]")
}, error = function(e) {
NA
# HR
})
p2 <- tryCatch({
round(summary(fit_now2)$coefficients[, "Pr(>|z|)"][1],3)
}, error = function(e) {
NA
# p
})
dt_return <- data.table(hos,n_p,n_p_e,ir_p,n_a,n_a_e,ir_a,HR,p,HR2,p2)
raw_table <- rbind(raw_table, dt_return) %>% arrange(hos)
### for meta analysis
HRlog <- tryCatch({
coef(fit_now)
}, error = function(e) {
NA
})
SE <- tryCatch({
sqrt(diag(fit_now$var))
}, error = function(e) {
NA
})
HRlog2 <- tryCatch({
coef(fit_now2)[1]
}, error = function(e) {
# NA
HRlog
})
SE2 <- tryCatch({
sqrt(diag(fit_now2$var))[1]
}, error = function(e) {
# NA
SE
})
meta_now <- data.table(hos, HRlog, SE, HRlog2, SE2)
meta <- rbind(meta, meta_now, fill = T)
}
return(list(raw_table, meta))
}
# step3 result
(main1 <- create_main_table(dt,'mace')[[1]])
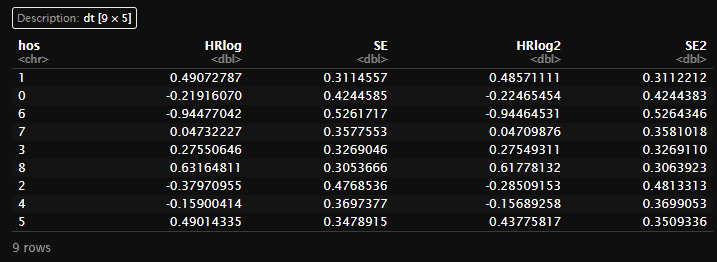
(meta1 <- create_main_table(dt,'mace')[[2]])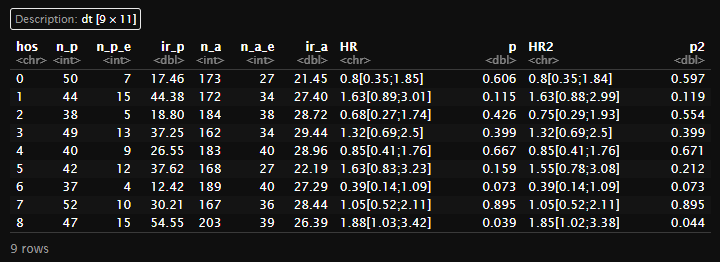
3. Meta analysis
- Meta analysis, Both of Fixed Effect and Random Effect
- Summary about FE, RE
https://vitalflux.com/fixed-vs-random-vs-mixed-effects-models-examples/
Fixed vs Random vs Mixed Effects Models - Examples - Data Analytics
Fixed effect, Random effect, Mixed effect models, Data Science, Machine Learning, Python, R, Tutorials, Tests, Interviews, News, AI
vitalflux.com
- Meta analysis function using metafor package
library(metafor)
# 1-2 meta analysis
table_meta <- function(dt){
# dt[is.na(HRlog2), HRlog2:= HRlog]
# dt[is.na(SE2), SE2:= SE]
# 1. Fixed Effect
res_FE <- rma(HRlog, sei=SE, data=dt, digits=2, method="FE")
res_FE2 <- rma(HRlog2, sei=SE2, data=dt, digits=2, method="FE")
# 2. Random Effect
res_RE <- rma(HRlog, sei=SE, data=dt, digits=2, method="ML")
res_RE2 <- rma(HRlog2, sei=SE2, data=dt, digits=2, method="ML")
# 3. output
meta_type <- c("FE","RE")
HR_FE <- paste0(round(exp(res_FE$b),2)," [",round(exp(res_FE$ci.lb),2),";",round(exp(res_FE$ci.ub),2),"]")
HR_FE2 <- paste0(round(exp(res_FE2$b),2)," [",round(exp(res_FE2$ci.lb),2),";",round(exp(res_FE2$ci.ub),2),"]")
HR_RE <- paste0(round(exp(res_RE$b),2)," [",round(exp(res_RE$ci.lb),2),";",round(exp(res_RE$ci.ub),2),"]")
HR_RE2 <- paste0(round(exp(res_RE2$b),2)," [",round(exp(res_RE2$ci.lb),2),";",round(exp(res_RE2$ci.ub),2),"]")
cat(HR_FE, HR_FE2, HR_RE, HR_RE2)
## pval > estimated / Not Heterogeneity
p_FE <- round(res_FE$pval,3)
p_FE2 <- round(res_FE2$pval,3)
p_RE <- round(res_RE$pval,3)
p_RE2 <- round(res_RE2$pval,3)
# 4. make dt
dt_meta <- data.table(hos=meta_type, HR=c(HR_FE,HR_RE), p=c(p_FE,p_RE), HR2=c(HR_FE2,HR_RE2), p2=c(p_FE2,p_RE2))
return(dt_meta)
}
(dt_meta<-table_meta(meta1))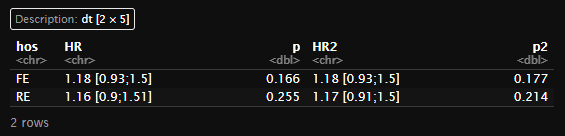
4. Hazard Ratio Table, meta anlaysis form for paper
Seo, Won-Woo, et al. "Impact of pitavastatin on new-onset diabetes mellitus compared to atorvastatin and rosuvastatin: a distributed network analysis of 10 real-world databases."
Cardiovascular Diabetology 21.1 (2022): 1-14.
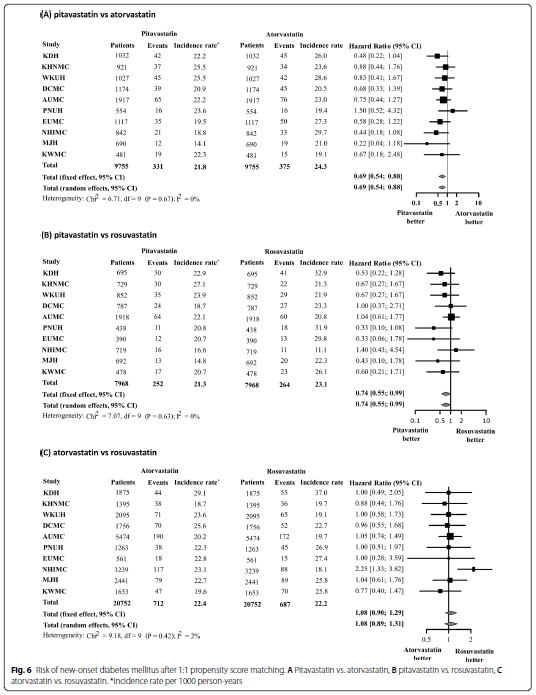
- Code to Mixing all outcomes
# 1-3 form_table
final_table <- function(main, meta, dt, outcome){
# 1-1 + 1-2 rbind and arrange by form
TOTAL <- data.table(
hos = "TOTAL",
n_p = nrow(dt[group=="Pravastatin"]),
n_p_e = nrow(dt[group=="Pravastatin"&get(outcome)==1]),
ir_p = round((nrow(dt[group=="Pravastatin"&get(outcome)==1])/sum(dt[group=="Pravastatin"]$event_tars_year))*1000,2),
n_a = nrow(dt[group=="Atorvastatin"]),
n_a_e = nrow(dt[group=="Atorvastatin"&get(outcome)==1]),
ir_a = round((nrow(dt[group=="Atorvastatin"&get(outcome)==1])/sum(dt[group=="Atorvastatin"]$event_tars_year))*1000,2)
)
dt_full <- rbind(main, TOTAL, meta, fill=T)
return(dt_full)
}
(f_main1 <- final_table(main1, dt_meta, dt, "mace"))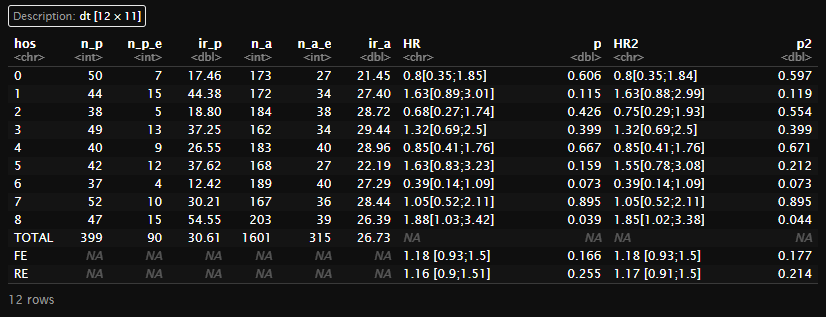
5. Forestplot
- Make a forestplot image
# 1. set your path
path<-"C:/Users/USER/Desktop/forestplot.png"
# 2. function to make a forestplot
mk_fp <- function(dt, path) {
library(forestplot)
library(ggplot2)
png(path,width=1750,height=600)
# make a header table
header <- data.table(
hos = c("","Institution"),
n_p = c("Pravastatin","Patient"),
n_p_e = c("", "Event"),
ir_p = c("","Incidence Rate"),
n_a = c("Atorvastatin","Patient"),
n_a_e = c("", "Event"),
ir_a = c("","Incidence Rate"),
HR = c("HR[95% CI]","Unadjusted"),
HR2 = c("","Adjusted"),
summary=T,
mean = NA,
lower = NA,
upper = NA
)
# transform according to forestplot form
base <- dt %>%
mutate(
mean = as.numeric(substr(HR2, start = 1, stop = regexpr("\\[", HR2) - 1)),
lower = as.numeric(substr(HR2, start = regexpr("\\[", HR2) + 1, stop = regexpr(";", HR2) - 1)),
upper = as.numeric(substr(HR2, start = regexpr(";", HR2) + 1, stop = regexpr("\\]", HR2) - 1)),
n_p = as.character(n_p),
n_p_e = as.character(n_p_e),
ir_p = as.character(ir_p),
n_a = as.character(n_a),
n_a_e = as.character(n_a_e),
ir_a = as.character(ir_a),
HR = ifelse(grepl("^NA", HR), "-", HR),
HR2 = ifelse(grepl("^NA", HR2), "-", HR2),
) %>%
select(-p) %>%
mutate(
summary = ifelse(hos %in% c("FE","RE"), T, NA)
) %>%
as_tibble()
source <- bind_rows(header, base)
plot<-source %>%
forestplot(mean=mean, lower=lower, upper=upper,
labeltext = c(hos, n_p, n_p_e, ir_p, n_a, n_a_e, ir_a, HR, HR2),
is.summary = summary,
clip = c(0.01, 5.5),
fontsize = 50,
txt_gp = fpTxtGp(label = gpar(cex = 1.2), ticks = gpar(cex = 1.2)),
xticks = c(0,0.5,1,1.5,2,3),
hrzl_lines = list("3" = gpar(lwd=1,lty = 1,columns=1:9)
# , "17" = gpar(lwd = 1, columns = 1:9, col = "#000044")
),
xlog = F,
col = fpColors(box = "royalblue",
line = "darkblue",
summary = "royalblue",
hrz_lines = "#444444"),
boxsize=0.2,
zero=1,
gap=unit(1,"mm"))
# save as png file
# png("forestplot.png",width=800,height=600)
print(plot)
dev.off()
return(plot)
}
mk_fp(f_main1, path)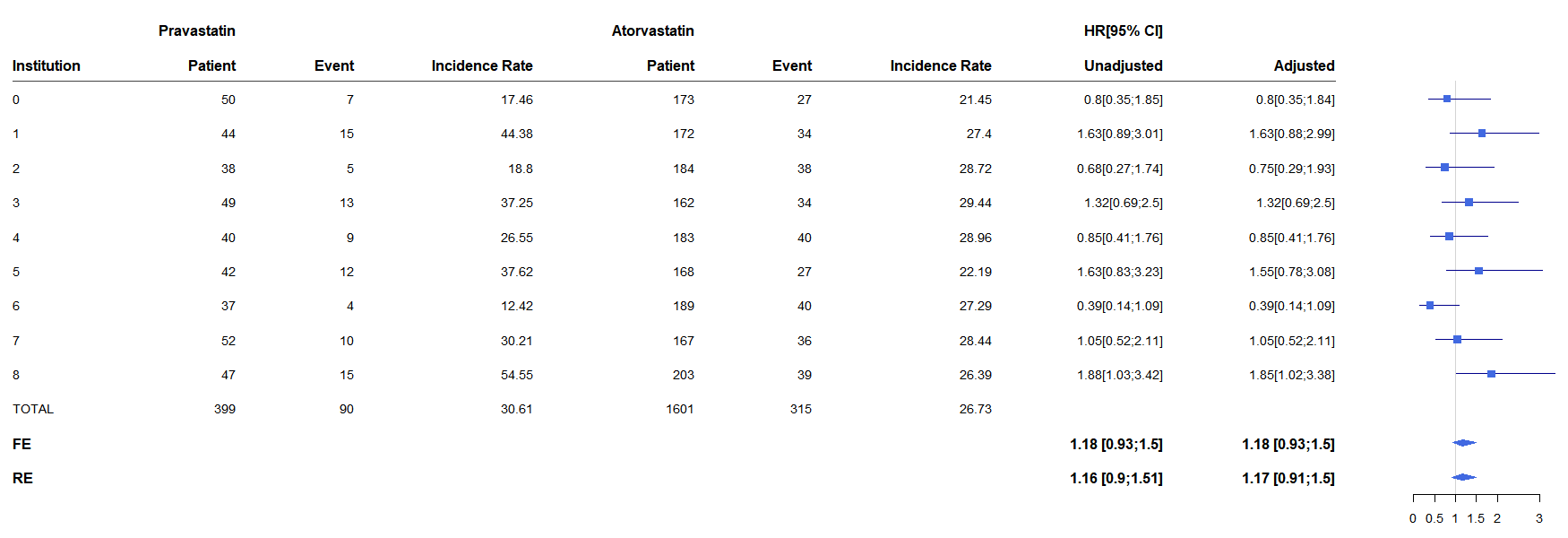
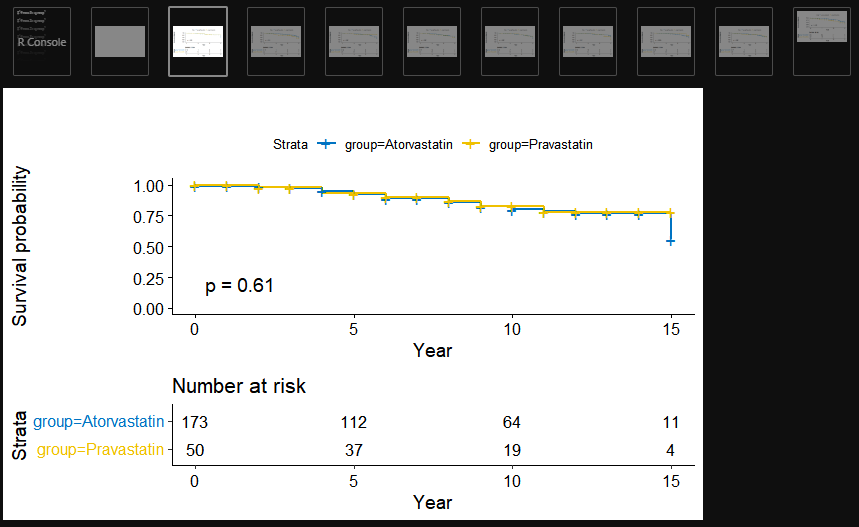
6. Kaplan-Meier
- Basic
library(survival)
library(survminer)
# 1. survfit model
fit <- survfit(Surv(event_tars_year, mace) ~ group, data = dt)
# 2. formula
km_total<-ggsurvplot(fit,data=dt,
risk.table=T, pval=T,
xlab="Year",
palette="jco",
# fun = "event", # event = cumulative event
# legend.title="Group",
# legend.labs=c("Atorvastatin","Pravastatin"),
tables.height = 0.35)
print(km_total)
# 2. save
ggexport(filename = "C:/Users/USER/Desktop/dskr_results/km/1. mace1/total_kaplan_meier.png",
plot = km_total, device="png")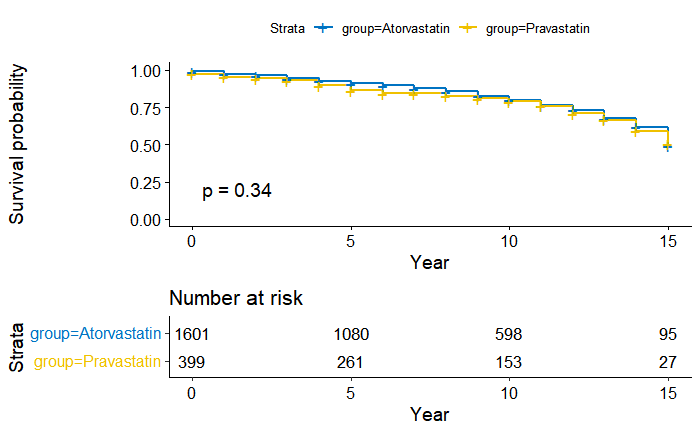
- Survival probability to Cumulative event
option fun = "event"
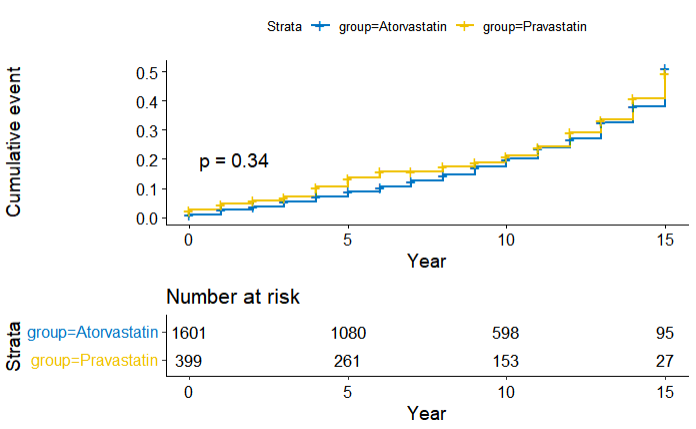
- Advanced, group_by
library(survival)
library(survminer)
# 1. survfit model
fit <- survfit(Surv(event_tars_year, mace) ~ group, data = dt)
# 2, group_by plot
gggb<-ggsurvplot_group_by(fit, data=dt, group.by = "from",
risk.table=T, pval=T,
xlab= "Year",
# fun = "event",
title="",
# legend.title="Group",
# legend.labs=c("Atorvastatin","Pravastatin"),
palette="jco",
tables.height=0.35)
print(gggb)
# print(gggb[[1]])
# 3. save as png file
hos_list <- unique(dt_meta$from)
for (i in 1:length(hos_list)){
km_name <- paste0("C:/Users/USER/Desktop/dskr_results/km/1. mace1/kaplan_meier_",hos_list[i],".png")
ggexport(filename=km_name, plot=gggb[[i]], device="png")
}